Ben Linn

PROJECT SUPERVISORS: Dr Sam Abraham, Associate Professor Darren Trott & Professor Roy Kirkwood , University of Adelaide
Benjamin Linn Looks at E coli
Benjamin Linn is in his final year of the University of Adelaide’s Doctor of Veterinary Medicine (DVM) program, having completed a Bachelor of Science (Veterinary Bioscience) in 2012.
Benjamin has always maintained a strong interest in pig production and from six years of age was even raising his own pigs. He also has a strong interest in pursuing a career in veterinary consultancy in pig production following graduation at the end of this year.
Throughout his studies, his achievements have been recognised with a number of awards, most recently the Chris Richards and Associates Prize in the Doctor of Veterinary Medicine program, the Australian Agricultural Scholarship, the Cowan Roseworthy Scholarship and the prestigious Audrey Abbie Veterinary Perpetual Prize.
In June, 2013 Benjamin commenced a Pork CRC supported project as his DVM-1 clinical research project, evaluating the prevalence of antibiotic resistance in commensal Escherichia coli isolated from pigs. Diarrhea caused by enterotoxigenic E. coli (ETEC) is an important cause of morbidity and mortality in neonatal and post-weaning piglets, manifesting as two major conditions: neonatal (ND) and post-weaning diarrhoea (PWD).
The enterotoxins secreted by ETEC cause hypersecretion of fluids into the small intestine, resulting in watery diarrhea and causing significant economic losses, decreased growth rates and treatment costs. Due to the highly virulent nature of ETEC strains, treatment tends to be aggressive, but resistance is emerging to medications commonly used to treat neonatal and PWD forms. In addition, treatment with antimicrobials may lead to colonisation of the gut with drug-resistant commensal E. coli.
The piggery chosen for this study had a history of outbreaks of diarrhoea, and the population of pigs chosen for study had been prophylactically treated with antimicrobials for prevention of enterotoxigenic E. coli (ETEC) infection.
Rectal swabs were taken from 105 randomly-sampled pigs from the piggery at various stages of production. Rectal swabs were obtained from pigs at one day of age (pre-treatment) and at entry to the weaner (four weeks of age), grower (eight weeks) and finisher (13 weeks) phases of production, and just prior to market (18 weeks). Samples were taken from two successive weaning batches, allowing for comparison between weaning groups. The swabs were then grown on antimicrobial-impregnated media specific for E. coli growth. Those that showed growth on this media underwent antimicrobial susceptibility testing using 18 antimicrobials commonly used in production animals and important to human medicine, as per Clinical Laboratory Standards Institute (CLSI) guidelines. These isolates were also tested for genetic relatedness and virulence genes, using Random Amplified Polymorphic DNA (RAPD) PCR and ETEC pathotyping multiplex PCR respectively. Those isolates resistant to antimicrobials of importance to human health were also screened to identify the genes responsible for the resistance.
In total, 86.7% of the swabs collected in this study exhibited multi-drug resistance. RAPD PCR demonstrated that the resistance isolates were heterogeneous in nature, however PCR pathotyping revealed the absence of any ETEC associated virulence genes, confirming that they were commensal E. coli.
This Pork CRC project identified that antimicrobial treatment of piglets results in persistence of multidrug-resistant commensal E. coli in the gut into the growing and finishing period. The resistant isolates were heterogeneous, indicating that more than one subtype of E. coli contains the resistance genes, which are most likely encoded on a plasmid.
The study indicated that the prophylactic use of antimicrobials in pig production systems may predispose pigs to the development of resistance amongst enteric commensal E. coli populations in pigs. Finding new ways to prevent/control ETEC infections in pigs will lead to less reliance on antimicrobial treatments.
The financial contributions of the Pork CRC and their support for this project are acknowledged. Thanks must also be given to Hui San Wong and Tahlia Mitchell for their considerable contributions to this project.
For more information contact Benjamin Linn, email benjamin.linn@student.adelaide.edu.au
First published in APN, April 2015
Anne Watt

Investigations into bacterial causes of disease in swine with emphasis on airborne pathogens
Pork CRC PhD candidate, University of Melbourne.
Supervisors: Dr Marc Marenda, Prof Glenn Browning and Assoc Prof Philip Markham
Watt’s Work All Up In The Air
Anne Watt recently entered the second year of her Pork CRC supported PhD at the University of Melbourne Faculty of Veterinary and Agricultural Sciences, as part of Pork CRC Project 2A-102, focusing on airborne pathogens in commercial settings.
She began the PhD after her Bachelor of Animal Science and Management with an honours year within the University of Melbourne’s Veterinary school. She obtained first-class honours for her work on the development of an indirect ELISA test for the detection of immune status of cattle to Mycoplasma bovis.
Anne’s research focuses on airborne pathogenic bacteria in commercial farming environments. Herd health and production performance is usually managed by monitoring environmental conditions, such as stocking density and ambient temperature and individual animal traits, such as weight gain and fertility rates. However, the impact of micro-organisms in commercial farming is difficult to measure because of the lack of simple, rapid methods for whole herd surveillance.
Managing infectious diseases in production animals currently requires collecting clinical or post-mortem specimens for isolation and identification of common pathogens by standard microbiological laboratory culture. This involves considerable time and expense that can be challenging for producers, particularly when samples contain mixed micro-organisms. Alternatively, detection of specific DNA sequences through Polymerase Chain Reaction (PCR) can rapidly identify a pathogen in such complex populations. Quantitative PCR assays with melt curve analyses are increasingly being used by diagnostic laboratories because of their excellent specificity and sensitivity.
Instead of individual clinical microbiology, sampling the atmospheric environment is a useful method for the detection of airborne materials in intensive animal production systems. Monitoring air quality on farms can provide non-biased, early warning data to improve livestock health status by preventing or controlling infectious diseases before they become a significant burden. Previously, there has been little work on the application of these methodologies to pig production. Now, through her current work, Anne has shown the benefits of such an approach.
She has already sampled air from two different commercial farm settings, covering eight different pig age groups and several housing systems. Air sampling is via a commercial air sampler, designed to assess biological contamination in different environments. This equipment captures micro-organisms present in a set volume of air, trapping them in a liquid media that can then be used for further analysis by PCR or culturing.
Following Anne’s first assessments, Actinobacillus pleuropneumoniae (APP) was chosen as an important pathogen present in pig farming and became the focus of her studies. APP, previously known as Haemophilus pleuropneumoniae, is the major cause of pneumonia in pig herds around the world. There are two biotypes and 15 known serotypes of APP, with the most virulent strains able to cause death within a matter of hours. If a herd contracts a highly virulent strain of APP, the damage to the herd can be severe, resulting in dramatic losses to the farmer.
With the idea that this air sampling method could be used to screen herds for the presence of APP in the environment, Anne has developed a quantitative PCR for detecting APP in air. From samples so far collected, she has detected low levels of APP in pigs aged eight to 16 weeks, indicating a sub-clinical infection within the herd, thus detecting the disease early enough to prevent production losses. The high resolution melt curve analysis also allowed identification of genetic differences within the sample, which was then confirmed though DNA sequencing. This technique allows for more specific identification of strains present and shows their relatedness.
This work on air sampling has naturally progressed into an attempt to develop a vaccine candidate for APP. Current vaccines have not successfully provided cross protection for all serotypes of APP, meaning identification of the strains present during an outbreak essential in order for protection to be provided. Anne’s vaccine work is focusing on producing a live-attenuated strain that has been restricted in its ability to utilise nutrients from its environment.
Anne’s research is driven by a desire to improve conditions in large scale commercial farming environments. Improving air sampling and diagnostic techniques can assist in rapidly identifying APP and lead to improved control measures. Future work on air sampling will look into developing quantitative PCR techniques for other important respiratory pathogens in the farming industry, with the aim to reduce losses in the industry though improved disease control.
Melbourne born and raised, Anne is still contemplating her career path post-PhD, but may consider a post-doc overseas, as she admits to a love of travel.
She also admits to a very real interest in farming conditions, especially differences in systems around the world and believes commercial farming is such a big industry.
Anne would particularly like to improve the welfare of animals, especially through improved disease control.
First published in APN Dec 2014
Lechelle van Breda

A comprehensive risk factor analysis of E.coli disease in the piggery environment
Pork CRC PhD candidate, University of Sydney.
Supervisors: Prof Michael Ward and Dr Om Dhungyel.
Finding E coli hard to resist
Lechelle van Breda is a Pork CRC funded student who has just completed the first year of her PhD as part of Pork CRC Project 2A-106. She commenced her PhD after completing Honours in marine science and a double degree in a Bachelor of Science/Bachelor of Creative Arts at the University of Wollongong, NSW.
Lechelle’s research focuses on pathogenic Escherichia coli (E. coli) disease that causes severe diarrhoea in weaner and suckling piglets. Weaning for the piglets is a very stressful time and can lead to the outbreak of gastrointestial diseases such as E. coli. Piglets are most susceptible to E. coli during weaning when they are experiencing changes in diet, gut flora and movement to a new environment. Pathogenic E. coli colonise the lower intestine where the bacteria produce enterotoxins, stimulating water and electrolyte loss, leading to dehydration and potentially death if left untreated. Clinical signs include diarrhoea or scours, dehydration, depression, lack of appetite and weight loss. It is a cause for substantial concern, as significant production losses are experienced, including reduced growth rates, high medication costs and high levels of mortality and morbidity.
Pulling triggers
According to Lechelle, we understand there are interactions between animals, the environment and humans, but we don’t understand why some farms experience this problem and others don’t. This is important for the Australian pig industry, as we don’t know what the key risk factors are that trigger an E. coli disease outbreak. We are also unaware of what and how often antibiotics are being administered to treat this disease and if there is any threat of antibiotic resistance to public health. Therefore, the aim of Lechelle’s project is to determine the high risk factors for E. coli disease in post-weaned piglets and ways to minimise those factors that could potentially contribute to a disease outbreak.
Farming faeces
A survey will assess and discuss key management and hygiene strategies with farmers to build a picture of everyday farm practices across several different commercial farms and draw key links between possible high risk factors and E. coli disease outbreaks. Based on this evidence, Lechelle anticipates that effective management strategies can be developed and implemented to reduce E. coli outbreaks, helping to minimise production costs. So far, 22 commercial piggeries in south-eastern Australia have been visited and faecal samples collected from pre and post-weaned piglets. These samples are being assessed to identify and determine the prevalence of different pathogenic E. coli strains present in Australia’s pig population. This knowledge is important and could assist future vaccine development.
Resistance patterns
The next step in this project is to assess the antimicrobial and disinfectant resistance patterns of E. coli in Australian pigs. As the threat of antibiotic resistance in public health is driving changes to the antibiotics available for livestock industries, alternatives to indiscriminate use of antimicrobials are needed. This project will determine if we are seeing any resistance to common antibiotics and if this can be reduced by simple changes to management practices, which can then provide an alternative long-term, sustainable solution to what is an emerging problem.
Pork CRC Program 2, ‘Herd Health Management’ aims to enhance animal health, while reducing antibiotic use in commercial pork production.
Contact Lechelle van Breda, email lechelle.vanbreda@sydney.edu.au
First printed in APN August 2014 issue
Bethany Bowring
Assays to measure gut health in order to identify risk factors and control strategies for E. coli scours in weaner pigs
Pork CRC Honours graduate, University of Western Sydney
Supervisors: Dr Alison Collins, Elizabeth Macarthur Agricultural Institute and Dr Ming Wu, University of Western Sydney
Healthy interest in piglet guts
Bethany Bowring received a Pork CRC Honours scholarship to study bacterial populations associated with piglet gut health in 2013, having completed her Bachelor of Science (Advanced Science) at the University of Western Sydney, graduating with Distinction in 2012.
Based at the NSW Department of Primary Industries’ Elizabeth Macarthur Agricultural Institute (EMAI) on the outskirts of Sydney, Bethany developed four different molecular assays to measure the numbers of beneficial and detrimental bacteria commonly found in piglet intestines. These assays were used to investigate relationships between groups of bacteria and scouring in piglets.
Assay aim
The ultimate aim of her research is to develop a ‘gut health’ assay for producers to evaluate diets and management practices to control scouring. The project is part of a bigger project funded by the Pork CRC to identify risk factors for scouring and alternatives to antibiotics for prevention and control of scouring. Following on from her earlier work, Bethany continues to investigate the impact of scouring on total numbers of bacteria and their diversity, working as a technical officer (part time) at EMAI.
Scouring options
The pig intestine contains an enormous number of bacteria (1014 or 100 million million) that are mostly beneficial to the pig, such as lactobacilli. They help digest nutrients and prevent infection by detrimental bacteria. Sometimes, however, these detrimental bacteria invade the gut and increase in number to cause disease. Scouring, caused by bacteria such as E.coli and Clostridia, is most common in the first two weeks of life and after weaning. At this time, piglet immune systems are not fully developed and piglets are adjusting to the stress of weaning and diet changes. Alterations in the balance of beneficial to detrimental bacteria within the gastrointestinal tract may contribute to scouring.
Left untreated, scouring is a significant cost to producers by reducing piglet growth and increasing mortality. Treatments including vaccination and antibiotics add to the cost of disease. However, some bacteria have developed resistance to the antibiotics used to control scouring, making them less effective. Many producers are keen to investigate alternatives to antibiotics to control scouring, including acidifiers in water and low protein diets. These management strategies may be evaluated by measuring the balance of beneficial and detrimental bacteria within the intestine.
CRC project
Dung samples from scouring and healthy piglets (before and after weaning) were submitted from seven different farms across Australia, along with information about antibiotic use. The number of beneficial (Lactobacilli) and detrimental bacteria (E.coli and Clostridia) were measured in each of the 121 samples. We expected to see higher numbers of detrimental bacteria in scouring pigs and higher numbers of beneficial bacteria in healthy pigs. However, these were not the results found, possibly because the numbers of beneficial and detrimental bacteria are not the same in dung and the intestine. We found that the number of E.coli in the gut did not correlate with the numbers found in dung from the same pig. However, Lactobacilli numbers in the gut did correlate with numbers in faeces. The complexity of the bacterial populations in the intestine may also have contributed to these unexpected results.
Routine destruction
Our project was also able to investigate the influence of antibiotics on bacterial numbers. Pigs fed with antibiotics had fewer Lactobacilli than non-medicated pigs. However, E. coli numbers in faeces were not significantly affected, suggesting that routine use of antibiotics may exacerbate scouring by destroying the beneficial bacteria instead of the bacteria causing scouring.
Industry impacts
Work is continuing to better understand which bacteria are important in gut health, including the impact of scouring on total number and diversity of bacteria. An assay that can measure the balance of beneficial and detrimental bacteria in scouring pigs could be used to evaluate risk factors and management tools. It is also hoped that the gut health assay would be a useful tool for producers wanting to monitor the impact of dietary changes aimed at preventing scouring, and treatment alternatives to antibiotics. Better control of scouring would reduce production losses and mortality and ultimately improve profitability. Finding alternatives to antibiotics to control scouring will alleviate consumer concern about drug resistant bacteria.
Contact Bethany Bowring, email bethany.bowring@dpi.nsw.gov.au
First printed in APN April 2014 issue
Rousset Palou

PhD topic – Maternal transfer of dietary essential oils and perinatal conditioning in piglets may help decrease antibiotic use , part of Pork CRC Project 2C-105
University of Queensland supervisor: Dr Eugeni Roura
Weaning is a critical period in development linked to changes in the social and physical environment, dietary regime, loss of the maternal protective passive immunity and particularly a low feed consumption during the first days. In addition, early weaning practices in commercial pig husbandry imply that the piglets have not completely developed their gastrointestinal tract, which further decreases their capacity to fight potential pathogens. Consequently, diarrhoea has a high incidence in the first weeks post weaning, generating a decrease in performance and, in severe cases, death. The principal microorganism causing post weaning diarrhoeas is E. coli. According to the Department of Agriculture, Fisheries and Forestry, piglet diarrhoea costs the Australian pig industry more than $7 million per year. For many years farmers were using antibiotics to prevent and treat the effects of post weaning pig diarrhoea. However, the efficacy of antibiotics is decreasing due to the emergence of bacterial resistance. Also, there is an increasing concern for potential human health implications associated with antibiotic resistance and their use in livestock production, therefore many countries are adopting restrictive policies for the use of antibiotic in feeds.
PhD research
Rousset’s PhD research topic at the University of Queensland is part of Pork CRC Subprogram 2C, which targets replacing antibiotics, designing effective integrated health strategies and addressing a novel approach to the use of plant essential oils (EOs). EOs are commonly used in the food industry as flavouring compounds, but they’ve also been reported as effective antibacterial agents (Burt and Reinders, 2002; Michiel et al., 2008), immunoprotectors (Halas et al., 2011) and performance enhancers (Li et al, 2012) in vitro. However, the antibacterial efficacy of EOs in vivo in weanling piglets is controversial since it lacks consistency. Most EOs are quickly absorbed and they may not reach the main site of proliferation of E. coli in sufficient amounts (Foley et al., 1986). Also, EOs seem to have a negative impact on feed intake in weanling pigs (as part of a ‘neophobic’ response very common in mammals), which may worsen the post weaning check. Our Pork CRC supported project addresses these two issues.
Firstly, there is evidence of the transfer of flavour compounds from the mother’s diet to offspring through milk and amniotic fluid in several mammals (Menella et al., 1995; Figueroa et al., 2011; Hepper et al., 2012; Oostindjer et al., 2011), which helps determine or condition postnatal preferences. We are studying products based in essential oils, with an effective antibacterial activity that, in addition, show good transference from sow feeds to maternal fluids. Exposing piglets to such essential oils in the peri-natal period may result in a pre-conditioning of the piglets to improve EO preference early in life which, in turn, will improve the post-weaning check.
Secondly, several technologies are being tested that may protect the EOs in the upper gastrointestinal tract and favour a release at the site needed to fight E. coli.
Essential evidence
Our preliminary data has already provided evidence that some of our selected EOs can be traced back from the sow feed to the milk. To our knowledge, the traceability of dietary EOs into milk samples analysed by GC-MS has never before been reported in sows. Our team is excited about that proof of concept and looks forward to upcoming developments.
Rousset’s profile
Born in Chile, Rousset Palou came to Australia nearly three years ago, after studying Veterinary Sciences at the University of Chile (Universidad de Chile). In Chile she worked in an EU Welfare Quality project, which was looking to improve animal welfare in slaughterhouses. She then decided to come to Australia for a better lifestyle for her family and for her husband’s job. About 10 months ago she started work as a Research Assistant at the University of Queensland in Dr Eugeni Roura’s group, preparing and organising pig trials related to taste and nutrition.
Contact details: Rousset Palou, email r.palou@uq.edu.au
First printed in APN July 2013 issue
Phil Markham
Pork CRC Post-Doc – Real time detection of airborne pathogens in the piggery – Pork CRC Project 2A-102
University of Melbourne Supervisor: Dr Marc Marenda
Phil Markham’s Pork CRC post-doc project, within the Herd Health Management Program and supervised at the University of Melbourne by Dr Marc Marenda, will help manage on-farm livestock health and performance by detecting a range of pig bacterial pathogens. It will also help to implement appropriate antimicrobial therapy, prevent or mitigate infectious disease outbreaks in pigs and contribute to the health of farm workers.
The technical focus of the research is to develop Polymerase Chain Reaction (PCR) assays to detect pathogens. These assays will be based on characterised genomic DNA sequences. They also aim to simultaneously detect DNA sequences that encode virulence determinants with pathogenic potential: this may be an antibiotic resistance gene(s) or genes encoding toxins causing disease.
The PCR assay has been developed based on all organisms possessing a unique DNA sequence that allows their classification down to the species level. The assay can then be used to show the presence or absence of a particular species. While this is useful in determining the presence or absence of a particular bacterial species, it is in some situations necessary to determine the level or bacterial load present and use this to predict herd health. With this in mind, as a proof of concept, a quantitative PCR has been developed to detect the level of Escherichia coli in a sample. They can then use the assay to quantify the number of genome copies and hence bacteria present, in a sample. The level of a particular organism can then be correlated to data on animal health and production results. Statistical analysis can then be conducted to investigate the parameters in pig production and presence of particular bacterial species.
As an example, virulent strains of enterotoxigenic E. coli (ETEC) possess genes that encode virulence determinants associated with diarrhea in neonatal pigs such as ‘heat labile and heat stable’ peptides. The PCR test will be used to detect such genes that also encode fimbriae responsible for attachment to the host. While possession of virulence determinants may cause disease, treatment using antibiotics might be considered a first line of defence. In this case the presence of the antimicrobial resistance gene repertoire is worthwhile knowing. Antibiotic resistance genes may be located chromosomally, extrachromosomally on a plasmid or on another mobile genetic element. They will use an air-sampling machine to collect a set volume of piggery shed air, with the air passed into a small volume of sterile diluent. The bacteria in the sample will be collected by to centrifugation, suspended in sterile diluent and serial dilutions prepared and plated onto selective agar media to enumerate bacterial numbers.
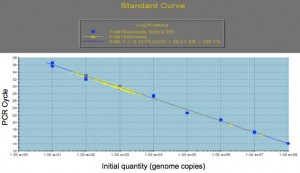
Separately, genomic DNA will also be extracted from the suspended bacterial sample and processed for quantitative PCR. Following PCR, the sample genome numbers will be determined by interpolation from a standard curve of known genome copies (see Fig 1).
Figure 1. Genome Copy Number vs PCR cycle
Air samples from a number of piggeries that represent ‘clean’ and ‘dirty’ environments will be tested by qPCR and analysed, along with husbandry data. This methodology allows a piggery to routinely collect an air sample for analysis and then respond accordingly, with husbandry changes to improve pig health.
After an undergraduate degree at Monash University, majoring in microbiology and immunology, Phil’s PhD was in avian mycoplasmology at the Veterinary Faculty at the University of Melbourne, where he has worked for 17 years and is now a Senior Research Fellow.
Contact details: Dr Phil Markham, email pmarkham@unimelb.edu.au
First printed in APN March 2013 Issue
Sarita Guy
PhD topic: ‘Genetic aspects of disease tolerance in pigs’ – University of Sydney
Project Supervisors: Assoc Prof Peter Thomson & Dr Susanne Hermesch
Sarita’s Pork CRC project will help develop new procedures in Australian pig breeding programs that incorporate disease tolerance in breeding decisions. She aims to formulate new genetic improvement strategies for selection of healthy pigs that are highly productive across a variety of environments. These new procedures will be based on new statistical models that use data from commercial farms. These data may include specific veterinary measurements and measures of pathogen load on-farm, which are currently being investigated in other Pork CRC research projects. While there is currently very limited information about breeding strategies for disease tolerance in livestock, the project has the potential to be a world-first for a number of research questions, thus boosting the relevance of Australian pig research and breeding internationally.
Healthy approach
Evidence suggests we can maintain the health and welfare of pigs without a significant decline in production, even when facing increasing disease and environmental challenges. Minimising veterinary intervention and medication use, while producing a more predictable and consistent pork, despite environmental fluctuations, matches the major themes of the CRC for High Integrity Australian Pork. To achieve this, she is investigating the host defence mechanisms of disease tolerance and disease resistance. These two mechanisms differ fundamentally in immune response and host-pathogen interaction and should not be used interchangeably. Resistance has been well characterised in pig breeding, with candidate genes found for Porcine Reproductive and Respiratory Syndrome (PRRS) resistance, for example. But there is little information describing disease tolerance. She emphasises there is a difference between the two mechanisms. Resistance refers to the mechanisms that actively reduce pathogen burden by inhibiting infection and reducing pathogen growth rate. A resistant pig will therefore successfully eliminate the pathogen. Tolerance mechanisms, however, aim to minimise and counteract damage from the pathogen. A tolerant pig is therefore more productive than a non-tolerant pig, despite increasing pathogen burden.
Tolerating differences
It can be argued that the main way tolerance differs from resistance is lack of interaction between host and pathogen. Both mechanisms have their advantages and disadvantages. Breeding for disease resistance may enable the eradication of a disease. This will not be achieved if pigs are selected for disease tolerance, as a tolerant pig may harbour the pathogen but not show symptoms.
Breeding for tolerance also does not decrease pathogen prevalence, as it allows pigs to be a source of infection for other susceptible pigs, thereby increasing transmission of infection. It follows that other management measures must be in place to control pathogen load if non-tolerant pigs are present on farm. On the other hand, resistance mechanisms directly exert pressure on the pathogen, prompting it to become more virulent, which may increase the need for the continual development of new medical products to eliminate the more virulent pathogens. These differences between the two mechanisms also imply that they require different procedures in order to incorporate them into pig breeding programs.
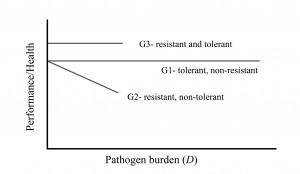
Graphic representation
The graph shows the difference between resistance and tolerance, but it should be noted that while this linear representation may be easily understood, in reality the relationship between performance and disease burden may not be linear. Also, please note this is an outline of the concept. Actual levels of performance or health of resistant versus tolerant pigs, for a given pathogen burden, will depend on the specifics of each situation.
A pig can be tolerant and non-resistant, resistant and non-tolerant, or tolerant and resistant, shown here as genotypes G1, G2 and G3, respectively. A fully tolerant pig is one whose performance does not decline, despite increasing pathogen burden. G1 and G3 are said to be more tolerant than G2 as performance or health is not affected by differences in pathogen burden for G1 and G3. A resistant pig is one that successfully eliminates the pathogen and so higher pathogenic burdens are not observed. G2 and G3 are more resistant than G1 because pathogen burden remains lower for G2 and G3.
She suggests that selecting for pigs that perform in a wide range of environments should incorporate not only tolerance to pathogenic challenges, but also any environmental challenges, which are often omitted when modeling and predicting resistance and tolerance.
Farm focus
To maximise benefit to the pork industry, she emphasises the use of readily available on-farm data, complemented by new data-logging technology which offers new opportunities. Such data may be used to construct an indirect measure (or proxy) of pathogen load, if, for example, a link is established between pathogen load and level of medication, or performance or survival rate of pigs. This information can be used for husbandry practices on-farm to improve health status of pigs. How such new data can be incorporated into pig breeding programs to select tolerant pigs, with improved health and productivity, is the focus of my research.
Contact details: Sarita Guy, Email Sarita.guy@sydney.edu.au
First printed in APN issue Oct 2012